Christian Parisod
PER 23 - 1.02
+41 26 300 8852
E-mail
What processes cause species properties to change over space and time?
What is the role of genomic (endogenous) and ecological (exogenous) constraints in shaping biodiversity?
To address these central questions, we use approaches from ecological genomics to assess stochastic, recombinational, selective and mutational drivers of the evolutionary ecology of plants at multiple scales. We combine approaches from genomics and ecology to characterize underpinnings of adaptation and speciation under changing environments.
As accumulating evidence points to significant genome dynamics at the origin of plant evolutionary-relevant variation, our interest is to integrate genomic and environmental drivers of diversification. We particularly address how gene duplication and transposition interact within and among species evolving across natural landscapes.
We rely on high-throughput genomics in natural and experimental populations to …
… infer population dynamics and adaptation using population/landscape genetics.
… assess genome dynamics with a focus on transposable elements and duplicated genes.
… integrate genomic and ecological changes using polyploid speciation as a model process.
Our current projects thus combine high-throughput sequencing technologies (DNA/RNA) and computational procedures with experimental and empirical field studies of alpine plant populations.
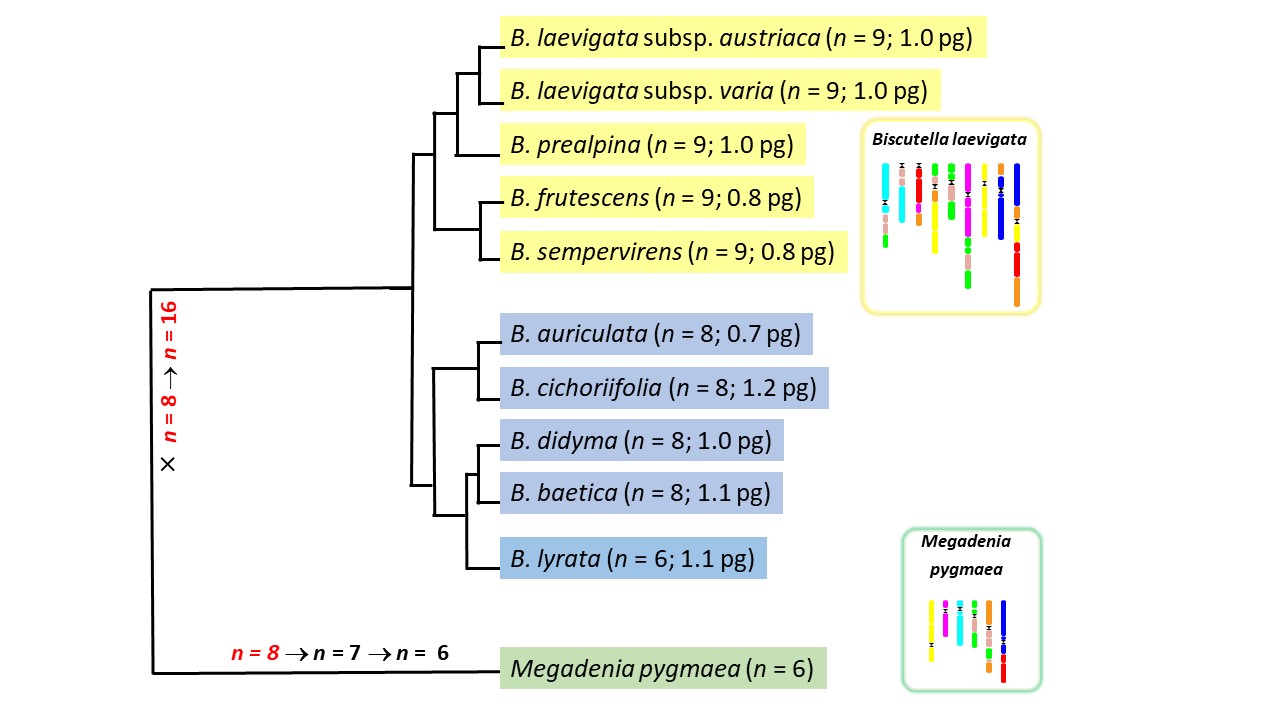
Colonization of alpine landscapes by Buckler Mustards (Biscutella laevigata; Brassicaceae) has been associated with recurrent whole genome duplications, dynamics of transposable elements and adaptive evolution of stress-responding genes. We focus on this mixed-ploidy model system to integrate genomic and ecological changes associated with autopolyploid speciation.
In particular, we currently use natural as well as experimental populations of diploids and autopolyploids to address underpinnings of their radiation under environmental constraints inherent to elevation and soil toxicity gradients.
We rely on whole-genome sequence data, transcriptomics and phenotyping to infer interactions of duplicated genes and transposable elements in response to contrasted habitats.
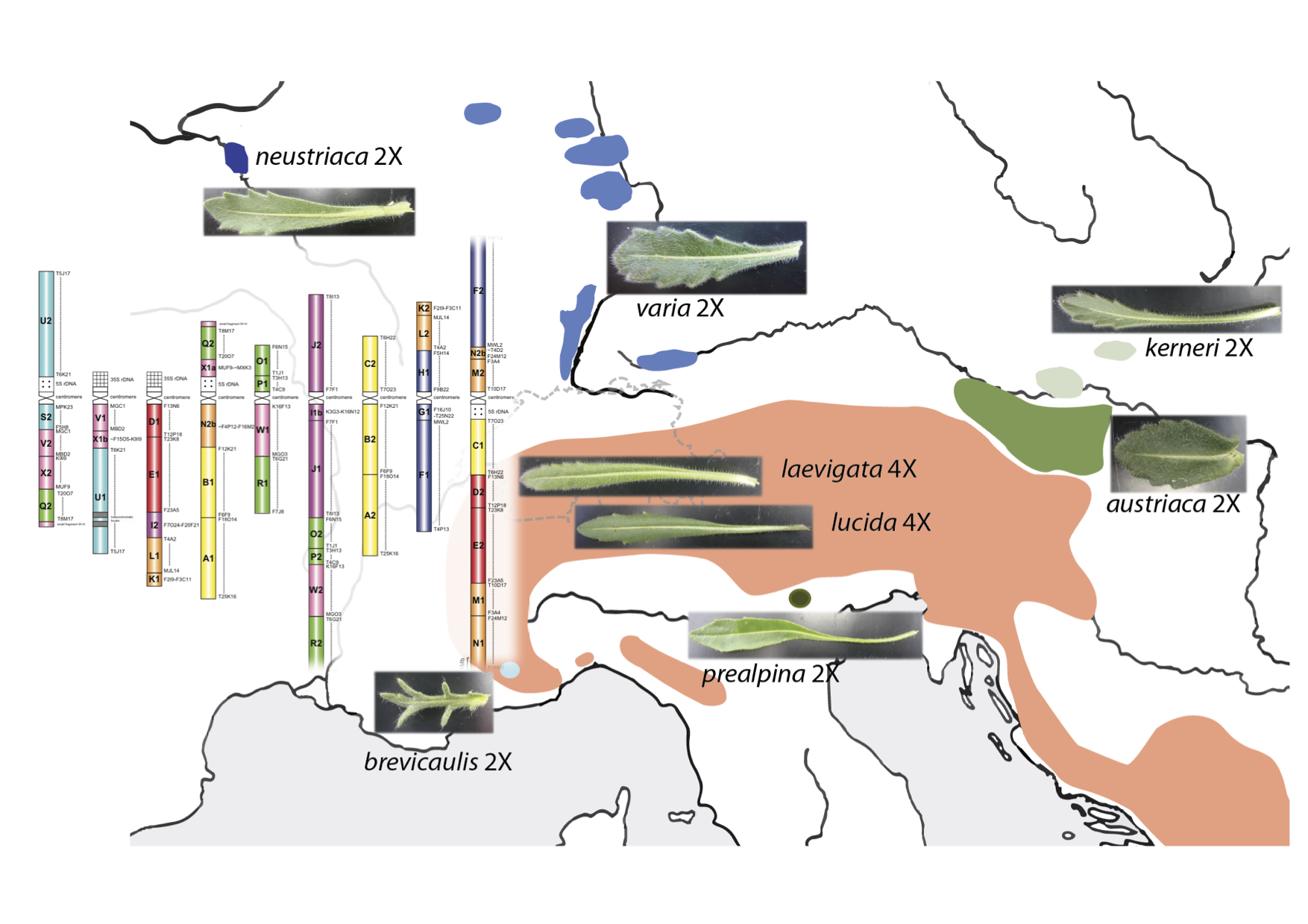
In collaboration with Martin Lysak (https://www.plantcytogenomics.org/), we sequence and assemble genomes from multiple Biscutella species sharing a common polyploidy event but differing in their chromosome numbers and/or genome sizes. Comparative genomics will shed light on the role of transposable elements, duplicated genes and structural variation on chromosomal divergence and dysploidy during species diversification across Mediterranean to Alpine environments.
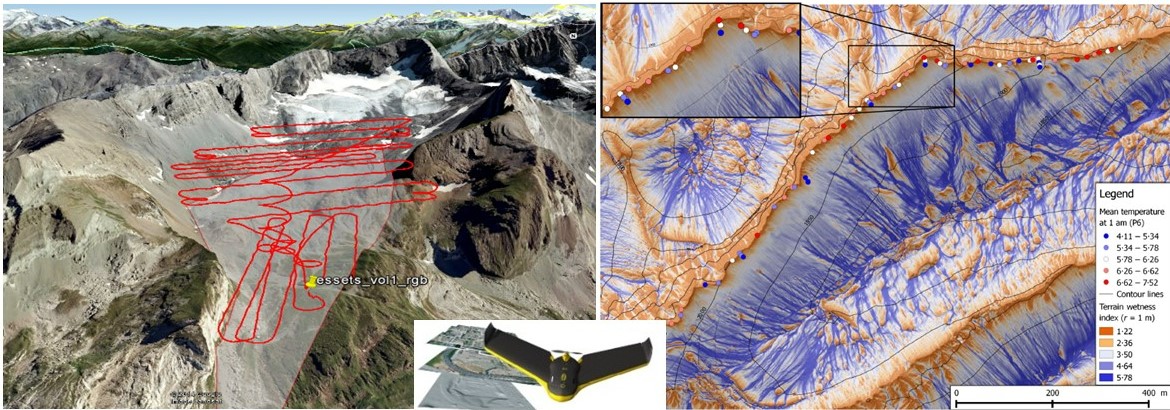
With the GeneScale consortium, we develop landscape genomics on alpine populations of Arabis alpina by associating genetic variation and environmental heterogeneity characterized at high resolution. Multi-scale decomposition tackles the effective scale of natural selection.
With Kim Gilbert, we investigate the evolutionary consequences of population expansion on genomic variation in Arabis alpina. Integrating simulations of complex demographic scenarios with sequence data from several hundred field samples, we address the impact of selection and particularly the accumulation of deleterious mutations during climate-induced range expansion.
We explore the evolutionary history and the population genetics of narrow endemics and endangered species. Through regular collaborations with gardens and other botanical institutions, we have been working on Pulmonaria Helvetica (https://doi.org/10.1093/aob/mcaa145), Draba nivalis (https://doi.org/10.1111/1755-0998.13280), Papaver occidentale (https://doi.org/10.1007/s00035-020-00238-3), … and we are still enthusiastic about exploring new models.

PER 23 - 1.02
+41 26 300 8852
E-mail